571p Assessment of the Stability and Biological Activity of Heme-Proteins Encapsulated In Hydrogel Membranes
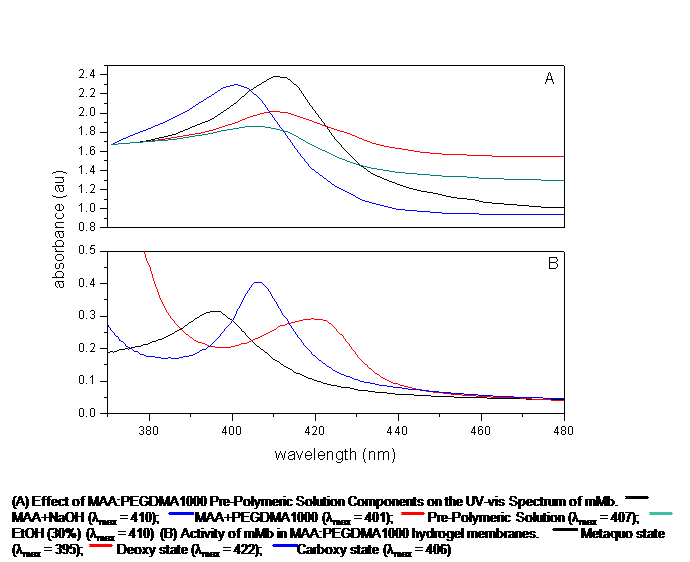
Proteins are biological macromolecules which have a unique spatial conformation. This spatial conformation can be affected by extremes in pH, temperature, and organic solvents. Once this 3D spatial conformation is compromised, the protein's biological stability and activity can be severely limited. This investigation focuses on the effects of pre-polymeric solution components on the behavior of proteins to be encapsulated by the entrapment technique in anionic, cationic, and neutral hydrogel membranes. Four proteins were utilized in this investigation: Equine skeletal muscle and heart myoglobin and Bovine and Porcine hemoglobin. Three hydrogel morphologies were examined: methacrylic acid-poly(ethylene glycol) dimethacrylate (n=1000), dimethylaminoethyl methacrylate-poly(ethylene glycol) dimethacrylate (n=1000), and poly(ethylene glycol) (200) monomethyl ether-methacrylate-poly(ethylene glycol) dimethacrylate (n=1000). The heme-proteins were put in contact with different ratios of the pre-polymeric solution components. UV-vis spectroscopy was utilized to monitor displacements in the Soret Bands to determine any changes in biological stability of the proteins in their metaquo, deoxy, and carboxy states. Optimized morphologies were then synthesized. The Soret bands of the proteins in all the pre-polymeric solutions were located at the expected wavelengths, 408nm and 406nm for myoglobins and hemoglobins, respectively. Upon polymerization, the Soret bands of the encapsulated proteins in ionic morphologies suffered blue shifts of 14nm, on average. Soret bands of the deoxy, and carboxy states were all blue-shifted by 16 and 18nm, respectively. The metaquo Soret bands of the four proteins encapsulated in the neutral morphology experienced a red shift of 4nm, whilst the deoxy and carboxy Soret bands presented a blue shift of 12nm and 5nm, respectively. These results suggest that the proteins undergo structural changes, most likely an iron-histidine bond breakage, due to the electrostatic interactions between the polymer and proteins. It appears that the neutral morphology did not cause such a remarkable alteration of the heme-proteins' spatial arrangement. Examination of the results also revealed a possible coexistence of metaquo/oxy and carboxy/oxy states of the proteins within the polymerized membranes.